Since its discovery in 2008, grapevine red blotch disease (GRBD) has negatively impacted the quality of wines due to reductions of sugar and color in the fruit. Its economic impact in the Western U.S. is estimated to range from $2,200 to $68,500 per vineyard depending on the growing region. Due to the presence of an insect vector capable of spreading the grapevine red blotch virus (GRBV), healthy grapevines can often become quickly infected during the growing season and symptoms can go unnoticed until the following season. Therefore, early detection of GRBV is even more crucial to preventing further transmission of the virus.
Symptoms of GRBD are often expressive in their characteristically red blotching patterns on leaves of red wine cultivars and likewise with yellow/yellow-white blotching on the leaves of white wine cultivars. However, symptoms on grapevines with established GRBV infections typically do not appear until after veraison. Consequently, molecular detection of GRBV can be critical for early determination of the infection status during early, non-symptomatic stages of infection.
Available GRBV Testing Strategies
There are multiple methods and strategies for diagnosing GRBD, including foliar symptom observation and monitoring, hyperspectral imaging, conventional polymerase chain reaction (PCR), quantitative PCR (qPCR), loop-mediated isothermal amplification (LAMP), plasmonic CRISPR and recombinase polymerase amplification (RPA). Among all these methods, PCR has remained the standard since 2014 due to its reliability, specificity and sensitivity; however, the PCR method creates technical, financial and infrastructure barriers for laymen due to the requirement for clean spaces, expensive instrumentation, complex troubleshooting and interpretation of results.
Other DNA-based methods such as LAMP and RPA, which are conducted at a stable reaction temperature throughout the procedure, do not require the same expensive equipment that PCR requires. The results from these two methods can be achieved much faster with reaction times as short as 20 or 30 minutes. In addition, LAMP and RPA are typically considered more sensitive to the DNA they target and less sensitive to impurities in the sample.

What is LAMP?
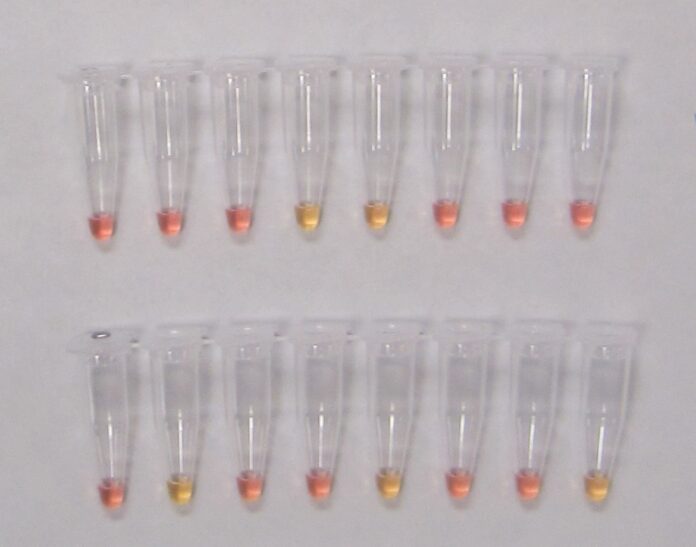
LAMP is a molecular tool used to detect DNA, commonly used as a diagnostic method for infectious diseases of plants and animals. Since its initial discovery by Notomi et al. (2000), the LAMP method has received much interest from private, academic and government sectors as well as from growers due to the low barriers to entry. In the past two decades, new formats for LAMP have been developed, making interpretation of results even more simplistic compared to the original method which had involved a gel electrophoresis system, additional chemicals and an advanced imaging system. These new formats allow the final interpretations to be done visually without any instrumentation. For example, the GRBV-positive reactions in some LAMP formats can create turbidity or cloudiness in the reaction tube, indicating GRBV was present in the sample, but more common is a change in color using pH indicators.
Recently, Romero Romero et al. (2019) published a LAMP method for the detection of GRBV along with a simplistic “pin-prick” DNA extraction method which consists of pricking leaf blades and petioles with a pipette tip and soaking them in water for 10 minutes to complete the extraction. Furthermore, these researchers paired this DNA extraction method with a colorimetric LAMP reagent that uses a pH indicator dye to determine whether the LAMP result was positive (yellow) or negative (pink) making interpretation faster and simpler.
We were interested in comparing its sensitivity and specificity to other more commonly used methods, such as PCR, qPCR and symptom monitoring.
The Experiment Design
We compared the four methods (LAMP, PCR, qPCR and visual symptom monitoring) at four different phenological time points per year for two years at a commercial vineyard in southern Oregon. We compared these methods using fully expanded, mature leaves sampled between berry set and harvest and using dormant shoot tissue during the winter. Both tissue types were collected at three different heights in the grapevine’s canopy: low-canopy (basal), mid-canopy and upper-canopy (apical). A tissue sample consisted of four leaves (one leaf from four shoots) or four dormant shoot segments and were collected for each canopy height and for each of the 40 vines used in this study. Vines were recorded for GRBD symptoms at the time of sample collection. Tissues were either subjected to a standard lab-based DNA extraction method and tested using PCR or qPCR or were subject to a simple, no-equipment-needed pin-prick DNA extraction method paired with LAMP.

What Was Discovered
In leaf samples, the accuracy of all methods was reduced when samples were taken from higher positions within the canopy. Therefore, we will present the remaining results of this experiment from the data collected from basal samples only since this is already standard practice for most virus testing.
The sensitivity, or ability to detect a positive sample, of all four methods differed significantly at all time points and canopy heights. At berry set and veraison, both PCR and qPCR successfully detected GRBV in 98% GRBV-infected samples across both years whereas LAMP could only detect GRBV in 49% and 78%, respectively, of the same GRBV-infected vines. Only 31% of these same GRBV-positive grapevines expressed symptoms during veraison. At harvest, qPCR detected 100%, PCR detected 98% and LAMP detected 96% of GRBV-infected samples. At this stage, 94% of grapevines were symptomatic. At dormancy, where there are no leaves to observe GRBD symptoms, 96% of the dormant shoots tested positive using PCR and LAMP, and 95% tested positive using qPCR. There was no statistically significant difference in false-positive rates (the percentage of samples incorrectly testing positive) between methods.
Due to the nature of this virus and its vector, some of our GRBV-negative vines became infected some time into the two-year experiment. Among the eight new infections observed, seven vines tested positive at our earliest sampling timepoint, berry set, by PCR and qPCR whereas LAMP only detected one of these vines at berry set and the other six thereafter. The eighth newly infected sample tested positive by all methods, but only at the harvest sampling.
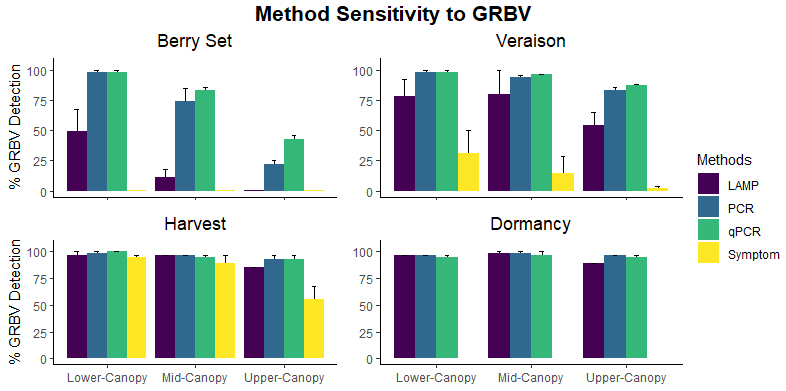
The conclusion from this experiment was the accuracy of these three DNA-based methods very much depends on the location of the sample in the canopy. Use of lower-canopy leaf samples later into season increased the accuracy of GRBV diagnosis and reduced the variability in detectability. It is evidenced that testing with LAMP for GRBV later in the season (e.g., near commercial harvest) can yield comparable results to more standard methods such as PCR or qPCR.
More cost-effective and simple methods such as pin-prick DNA extraction and LAMP can offer a more accessible approach compared to external testing or the barriers and complexities of performing PCR in-house. While PCR and qPCR testing of GRBV remains the more accurate method when testing until veraison, this experiment suggests LAMP can serve as a useful tool for those who may be seeking alternatives to PCR testing. LAMP may be of interest for those wanting to test more routinely, closer to commercial maturity or during dormancy when foliar symptoms are absent.